Citing
Yang J, Hou L, Liu K M, et al. ChemGenerator: a web server for generating potential ligands for specific targets[J]. Briefings in Bioinformatics, 2020.
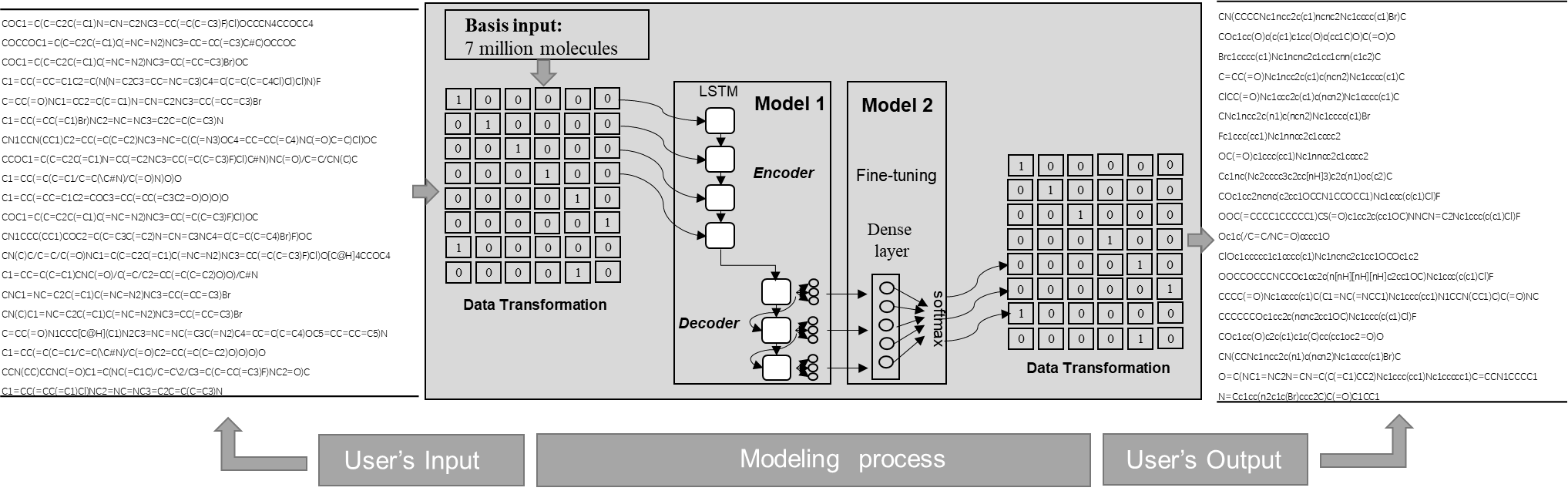
This web server is a novel SMILES strings prediction generator based on LSTM neural networks.
The input file for ChemGenerator is a list of SMILES strings. In many cases different toolkits use different algorithms for SMILES notation process. For this research, the training SMILES strings are consistent with the algorithm in PubChem database. So the results would be more credible if the input SMILES strings are canonicalized with PubChem database.
Output File
The first line of the output file consists of 2 values, one is value of Loss, and the other is Accuracy.
The rest of lines are newly generated SMILES strings on specific target.
A Brief Introduction to ChemGenerator
Chemical compounds can be expressed as a simplified language by Simplified Molecular-Input Line-Entry System (SMILES). ChemGenerator is a unique SMILES strings generator based on Long Short-Term Memory (LSTM) networks. This server is constructed with two solid models: the basic model and fine-tuning model.
By pretraining nearly 7 million molecular SMILES strings, the basic model secures that the 98% generated SMILES strings are valid molecules. The fine-tuning model focuses on target-guided molecule generation by transfer learning of the basic model.
In the webserver, you can input SMILES strings active toward one specific target, which will be treated as input sets for your fine-tuning model. As you know, adequate training is essential for modeling. To achieve a satisfying performance, we suggest you input relevant strings as more as possible (e.g., more than 1,000 strings), even though smaller datasets could also be trained. The supported input file formats are .csv and .smi, while file size should be less than 10MB. Generated molecules relevant to your specific target will be sent to you by email in three days. Meanwhile, we will provide you model evaluation results based on your dataset and appropriate user instruction. Due to the nature of purely academic work, the whole analysis is free of charge.